Project 10
A project that uses Python with ArcPy library
Automating development of weed prescription map using Arcpy and Python
Rai, N & Flores, P
Background
This algorithm imports ArcPy, and Rasterio packages in Python (Jupyter notebook) to create a weed prescription map. A feature to calculate Excess Green and four different vegetation indices (VIs), namely, Normalised Difference Vegetation Index (NDVI), Normalized Difference Red Edge Index (NDRE), Soil Adjusted Vegetation Index (SAVI), and Optimized Soil Adjusted Vegetation Index (OSAVI) can also be calculated and exported to local drive using the same script. The script accepts a multispectral or an RGB imagery. Based on band count, it calculates the required indices. After indices calculation, it also performs image sharpening and thresholding, thereby converting the whole image into a binary one. After that it converts objects within the image to polygons and performs weed identification while creating a weed map using Fishnet Grid technique. Full Python code to automate the process can be found here.
Prerequisites:
- RGB or multispectral imagery should be provided by the user with proper understanding on band information and geospatial location information.
- Specify a directory with an empty folder before this scripts starts exporting all the indices (output images) or processed images.
- Shapefile (a polygon line drawn over the crop plants) should be provided by the user.
Limitation of this algorithm:
- Weeds won’t get detected or identified if they are present in-between crop rows.
Code breakdown
-
Importing packages
This section imports various Python packages that are used throughout the script, including numpy, arcpy, rasterio, gdal, matplotlib, and others. These packages provide functionality for working with spatial data, raster manipulation, and plotting.
# This script creates a weed map using several ArcGIS tools and a bit of Image Processing.
# Original workflow in ArcGIS was developed by Dr. J. Paulo Flores (Assistant Prof. at NDSU)
# Automation using Python scripting was developed by Nitin Rai (PhD Student)
# Agricultural Engineering, Precision Agriculture
# North Dakota State University, USA
#################### Script starts here ########################################################################
################################################################################################################
# Importing packages
import numpy as np
import arcpy # from ArcGIS
import os
import sys
from arcpy.ia import * # Image Analyst
from arcpy.sa import * # Spatial Analyst
from arcpy import env
from arcpy.sa import Raster, Float
import rasterio # Working with Raster Dataset
import gdal # Geospatial Data Abstraction Library
import matplotlib.pyplot as plt
-
Reading imagery from the directory
Here, a raster image file (file_name) is specified, and its bands are extracted using the arcpy.ListRasters() function.
RGB_imagery = r"Your Directory/Sunf_AOI_Lab7_2021.tif"
raster = rasterio.open(RGB_imagery)
bands = [Raster(os.path.join(RGB_imagery, b))
for b in arcpy.ListRasters()]
-
Band count check in the imported imagery
Here the operation is dependent on band count. If the band count is equal to 5 then the script assumes it’s a 5-band imagery and starts calculating all the indices. More indices can be added to the workflow. Whereas, if the band count is greater than 5, then an error message is displayed asking user to enter an imagery which is either 3-band or 5-band. If the band count is equal equal to 3 or 4, then Excess Green is automatically calculated and stored to the HDD. As in array function, the index starts from 0 onwards. For a multispectral imagery (RedEdge MicaSense), 0 = Blue band, 1 = green, 2 = red, 3 = RedEdge, 4 = NIR
if band_count > 5:
print("Input Imagery should be 3-band RGB Imagery or 5-band Multispectral Imagery")
elif band_count == 5:
path = os.path.dirname(imagery)
base1 = os.path.basename(imagery)
base = os.path.splitext(base1)[0]
ndvi_filename = base + "_NDVI.tif"
ndre_filename = base + "_NDRE.tif"
savi_filename = base + "_SAVI.tif"
osavi_filename = base + "_OSAVI.tif"
-
Band calculation
Calculating the number of bands based on the formulae. You can add your own VI and extract information accordingly.
ndvi = (Float(bands[4]) - bands[2]) / (Float(bands[4]) + bands[2])
ndvi.save(ndvi_filename)
print ("NDVI successfully calculated")
ndre = (Float(bands[4]) - bands[3]) / (Float(bands[4]) + bands[3])
ndre.save(ndre_filename)
print ("NDRE successfully calculated")
savi = 1.5 * ((Float(bands[4]) - bands[2]) / (Float(bands[4]) + bands[2] + 0.5))
savi.save(savi_filename)
print ("SAVI successfully calculated")
osavi = 1.16 * ((Float(bands[4]) - bands[2]) / (Float(bands[4]) + bands[2] + 0.16))
osavi.save(osavi_filename)
print ("OSAVI successfully calculated")
else:
path = os.path.dirname(RGB_imagery)
base1 = os.path.basename(RGB_imagery)
base = os.path.splitext(base1)[0]
ExGreen_filename = base + "_ExcessGreen.tif"
total_bands = (Float(bands[0] + bands[1] + bands[2]))
red_band = (Float(bands[2]) / total_bands)
green_band = (Float(bands[1]) / total_bands)
blue_band = (Float(bands[0]) / total_bands)
ExGreen = (2 * green_band - red_band - blue_band)
ExGreen.save(ExGreen_filename)
arcpy.env.workspace = path
print ("You have successfully exported the Excess Green.tif file")
arcpy.BuildPyramids_management(ExGreen_filename, "", "NONE", "BILINEAR","", "", "")
print ("Excess Green pyramids successfully calculated")
-
Image sharpening
Sharpening the image so the objects (crops & weeds) details are highlighted. The Excess Green image is sharpened using convolution, and then a binary raster is created by applying a threshold.
sharpen_ExGreen = arcpy.ia.Convolution(ExGreen, 20)
sharpen_ExGreen.save(r'Your Directory/ExGreen_Sharpened.tif')
print("You have successfully sharpened the Excess Green.tif file!")
# Converting the above sharpened image into binary image, thresholding technique is applied here
# converting the imagery into 0 and 1.
binary_raster = arcpy.ia.Threshold(sharpen_ExGreen)
binary_raster.save(r"Your Directory/ThresholdSharpen.tif")
print("You have successfully perfromed Imagery Thresholding!")
-
Raster to Polygon conversion
The binary raster is converted to polygons, where pixels with value 1 become polygons.
inRaster = "ThresholdSharpen.tif" outPolygons = r"Your Directory/RasterToPolygonConvert.shp" arcpy.RasterToPolygon_conversion(inRaster, outPolygons, "NO_SIMPLIFY", field) -
Shapefile selection and buffering
Selected features (gridcode > 0) are saved to a new shapefile, and a line shapefile provided by the user is buffered.
selectedAttributes = arcpy.SelectLayerByAttribute_management(polygonAttached, "NEW_SELECTION", '"gridcode" > 0')
Buffered_CropsLine = arcpy.Buffer_analysis(cropsline, rowsBuffered, distanceField, sideType, endType, dissolve)
-
Polygon erasing and Fishnet creation
The script erases features from one polygon layer using another and creates a fishnet grid.
ErasedLayer = r'Your Directory/ErasedPolygonLayerFinal.shp'
arcpy.Erase_analysis(polygon, eraseFeature, ErasedLayer)
outFeatureClass = "fishnet_10x10.shp"
arcpy.CreateFishnet_management(outFeatureClass, origin_coordinate, yAxisCoordinate, CellWidth,
CellHeight, numRows, numCols, oppositeCorner,
labels, templateExtent, geometryType)
-
Field addition and spatial selection
A field is added to the fishnet, and a spatial selection is performed to select features that intersect with the fishnet.
AddedField = arcpy.AddField_management(inFeatures, addfield, "SHORT", fieldPrecision, field_is_nullable="NULLABLE")
LocationSelection = arcpy.SelectLayerByLocation_management(ErasedLayer, 'INTERSECT', outFeatureClass)
-
Copying selected features
The selected features are copied to a new shapefile.
arcpy.CopyFeatures_management(LocationSelection, 'IntersectFishnetwithErasedLayer')
Final output of the developed weed prescription map:
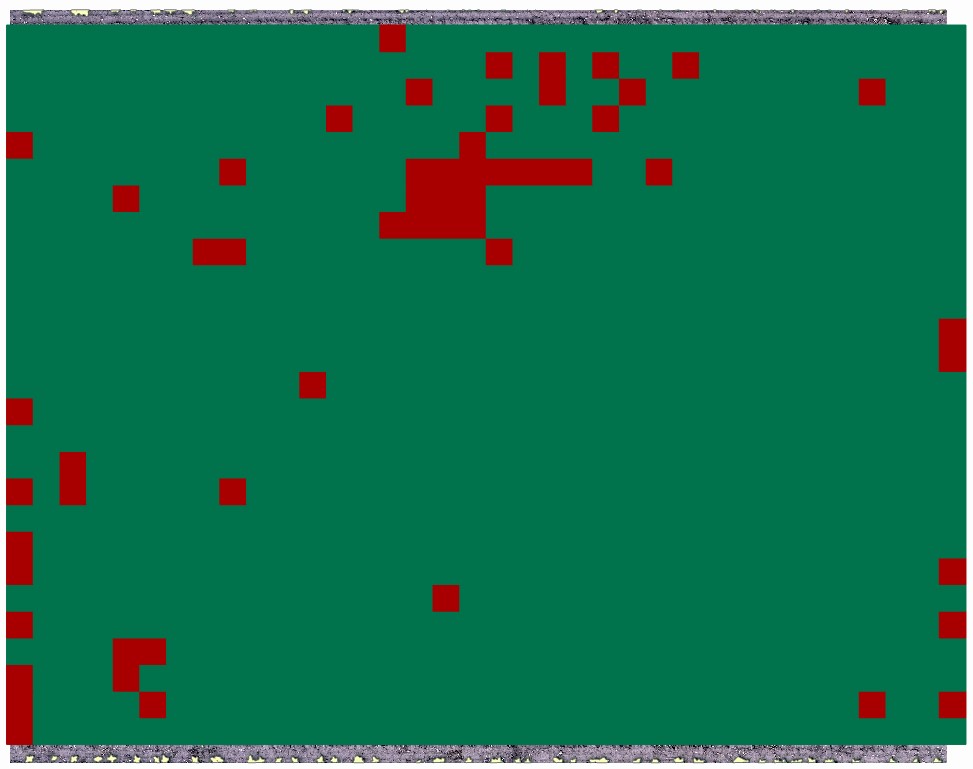
Credits:
- Original workflow in ArcGIS Pro was developed by Dr. J. Paulo Flores (Assistant Prof., Department of Ag. & Biosystems Engineering)
- Automation using Python scripting was developed by Nitin Rai